Welcome to
Intronserter
- a tool for built to aid the
design
of
intron-containing
optimized
transgenes
for enhanced nuclear expression.
Intronserter was developed primarily for
Chlamydomonas reinhardtii,
where systematic intron spreading into transgenes is necessary
to enable robust expression from the nuclear genome.
However, the Intronserter platform is fully customizable to any other
target organism by enabling user-defined inputs such as codon tables
and intron sequences.
By default, this tool provides full codon optimization and intron
spreading to encourage design of optimized transgenes for maximal
expression from the eukaryotic algal nuclear genome.
The strategy has been successfully applied in
C. reinhardtii
with numerous examples in literature:
Yunus, I.S., Wichmann, J., Wördenweber, R., Lauersen, K.J., Kruse, O., Jones, P.R., 2018.
Synthetic metabolic pathways for photobiological conversion of CO2 into hydrocarbon fuel.
Metab. Eng. 49, 201?211. doi:10.1016/j.ymben.2018.08.008
Baier, T., Kros, D., Feiner, R.C., Lauersen, K.J., Müller, K.M., Kruse, O., 2018.
Engineered Fusion Proteins for Efficient Protein Secretion and Purification of a
Human Growth Factor from the Green Microalga
Chlamydomonas reinhardtii.
ACS Synth. Biol. 7, 2547?2557. doi:10.1021/acssynbio.8b00226
Baier, T., Wichmann, J., Kruse, O., Lauersen, K.J., 2018.
Intron-containing algal transgenes mediate efficient recombinant gene
expression in the green microalga
Chlamydomonas reinhardtii.
Nucleic Acids Res. 46, 6909?6919. doi:10.1093/nar/gky532
Lauersen, K. J., Wichmann, J., Baier, T., Kampranis, S. C., Pateraki, I., Møller, B. L., Kruse, O. (2018).
Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from
Chlamydomonas reinhardtii.
Metablic Engineering. doi:10.1016/j.ymben.2018.07.005
Wichmann, J., Baier, T., Wentnagel, E., Lauersen, K. J., Kruse, O. (2017).
Tailored carbon partitioning for phototrophic production of (E)-?-bisabolene from the green microalga
Chlamydomonas reinhardtii.
Metabolic Engineering 45, 211-222. doi:10.1016/j.ymben.2017.12.010
Lauersen K. J., Baier T., Wichmann J., Wördenweber R., Hübner W., Huser T., Kruse O. (2016).
Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga
Chlamydomonas reinhardtii.
Metabolic Engineering 38, 331-343. doi:10.1016/j.ymben.2016.07.013.
The intron spreading strategy used for engineering nuclear transgene expression with
C. reinhardtii
may also enhance transgene expression in other species, especially in other green algae.
Therefore, Intronserter was designed so that all input parameters can be adjusted
to fit any desired host.
However, the enhancing effect of regulatory introns should be investigated first in
other species, and the strategy outlined in Baier et al, 2018, might assist in this process.
Intronserter starts from an input amino acid sequence, and yields its optimized (codon-optimized and intron-enriched) DNA sequence counterpart,
which is ready for gene synthesis, as outlined below:
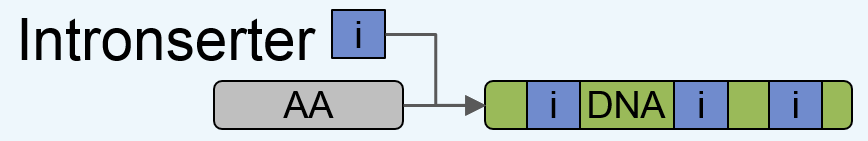 Figure 1: Outline of Intronserter.
Figure 1: Outline of Intronserter.
Towards this goal, Intronserter performs four steps:
1. Back translation of the Amino Acid (AA) sequence using the most frequent codon for AA.
2. Removal of restriction digest sites, so that, by default, restriction sites are avoided which are used in pOptimized and MoClo vector kits.
3. Enriching the cDNA sequence with introns to maximize expression
(here, the regulatory intron "rbcS2 intron 1" is used, as it has been shown to boost transgene expression
in C. reinhardtii (see Baier et al, 2018).
4. Optional fine tuning of the resulting DNA sequence to add for instance a linker peptide for fusions.
Step 3, the process of intron insertion, can be further dissected into three sub-steps:
3.1 Determination of intron positions:
3.1.1 Determination of putative intron insertion positions, based on the set of the four
parameters "start, target, max and end".
3.1.2. At these position, the nucleotide pair, e.g. "G^G", where the intron is inserted, is searched.
3.2. The supplied intron DNA sequence is inserted between the two bases given by nucleotide pair, i. e., between G^G.
3.3. As a optional last step, the last intron can be substituted for another intron sequence,
in order to maximize transgene expression in certain scenarios.
This process is outlined below:
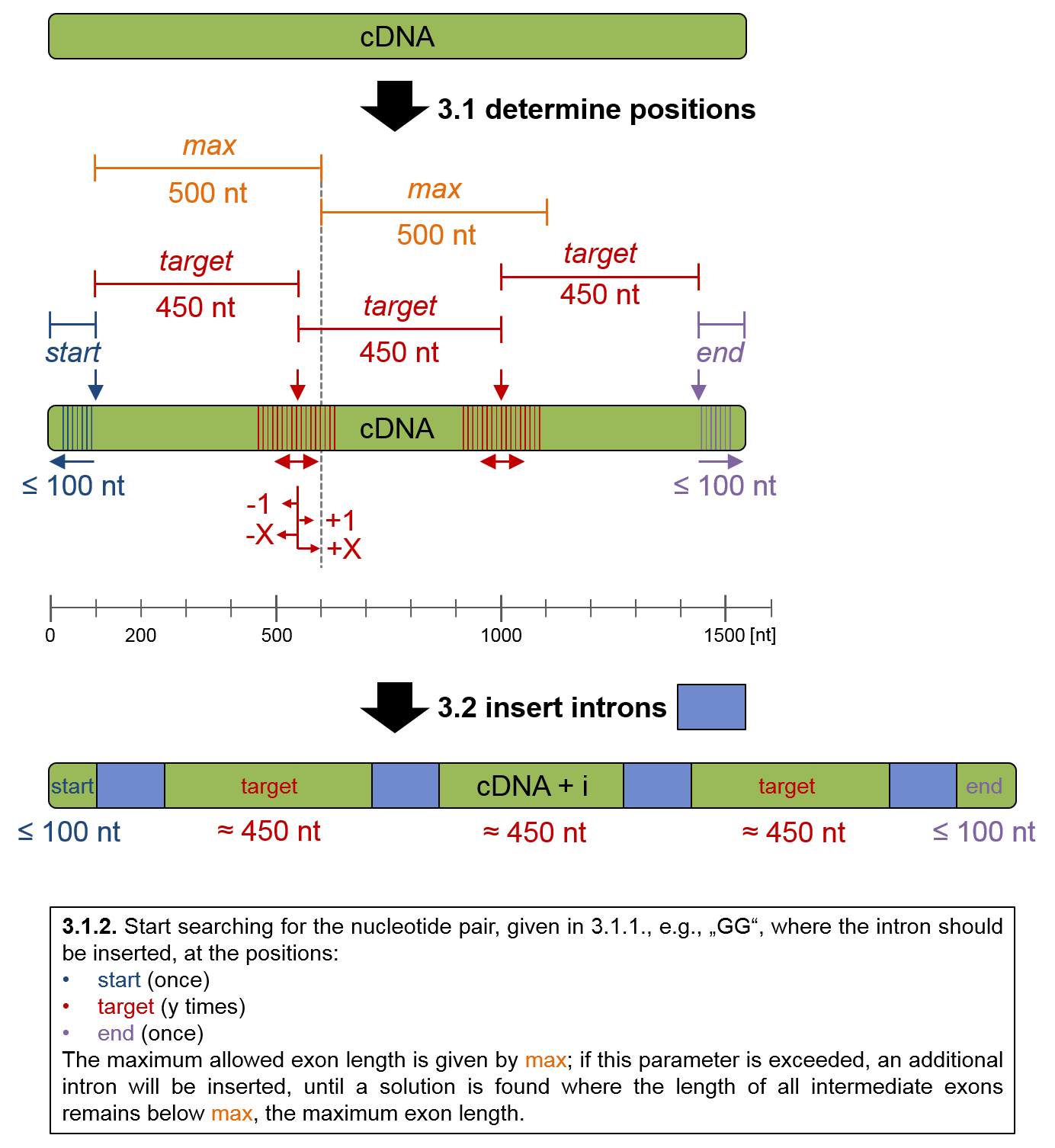 Figure 2: Intron insertion process.
Figure 2: Intron insertion process.
Step 3.3., the optional substitution of the last intron for "rbcS2 intron 2", is recommended in
C-terminal fusions in the pOptimized vector toolkit (see Baier et al, 2018), and for other vectors:
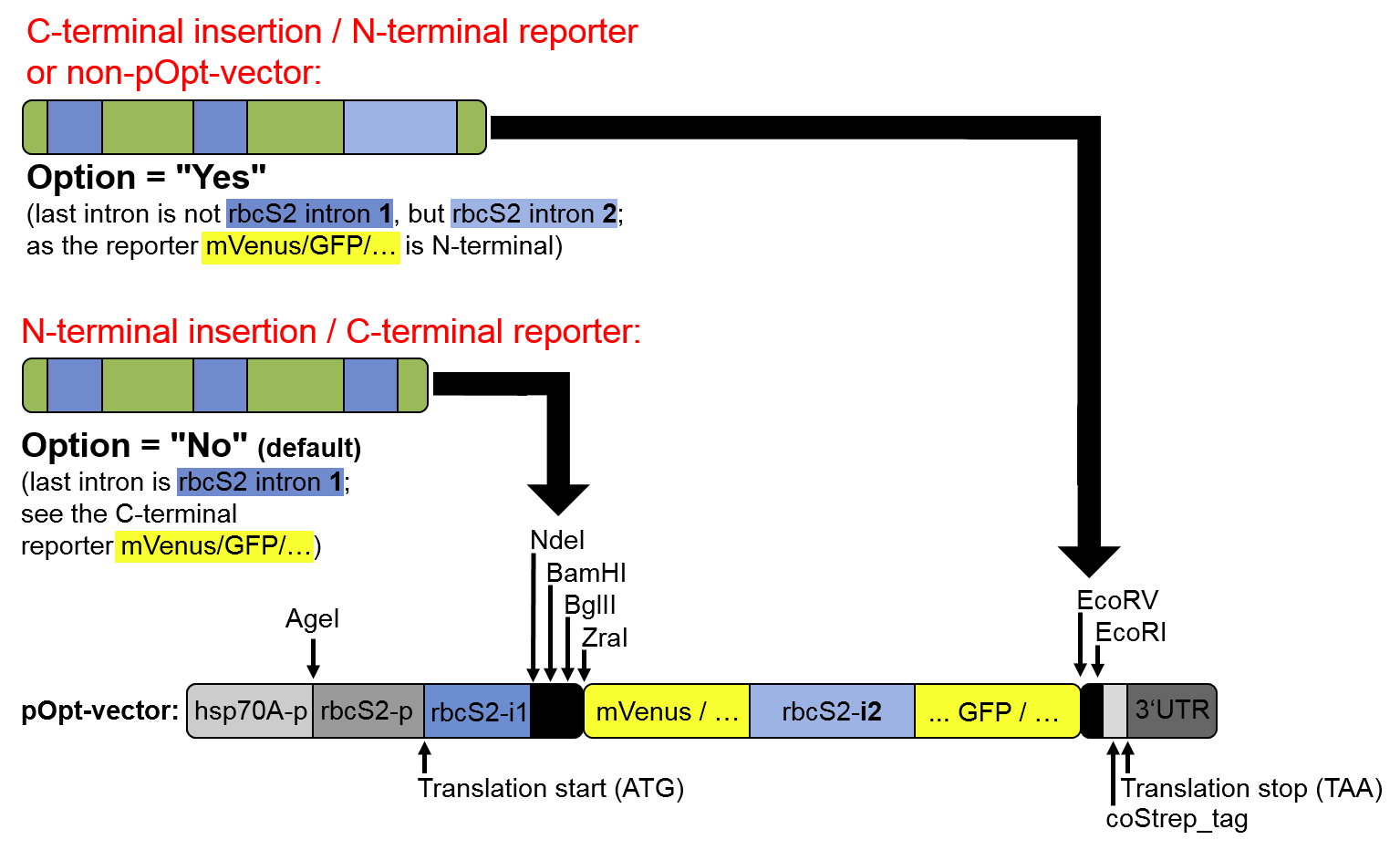 Figure 3: Scenarios in which substitution of the last intron for
rbcS2i2,
instead of using
rbcS2i1 only, is recommended.
Figure 3: Scenarios in which substitution of the last intron for
rbcS2i2,
instead of using
rbcS2i1 only, is recommended.
Step 4, the optional fine tuning of the optimized sequence, has the following ordering:
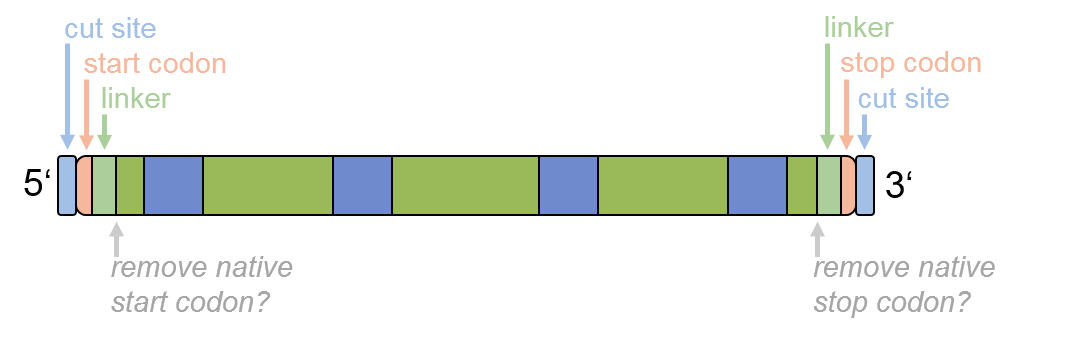 Figure 4: Ordering of the options for sequence fine tuning.
Figure 4: Ordering of the options for sequence fine tuning.
Below, the individual parameters are explained in more detail.
In-/Output values
INPUT :: Amino Acid sequenceAmino Acid sequence in FASTA format that is converted by Intronserter to
its optimized DNA counterpart. Uploaded file must be a .txt file type, encapsulated file types like .doc or
.rtf are not supported. For an example, click on the corresponding
button below.
INPUT :: User defined codon usage table User defined codon usage table, supplied in the standard codon usage table format /
in NCBI's standard genetic code definition,
found for instance at
Kazusa
or
HIVE-CUT
. Uploaded file must be a text file, encapsulated file types like *.doc or *.rtf are not supported. For an example, click on the corresponding button below.
INPUT :: DNA sequenceDNA sequence in FASTA format into which introns are inserted by
Intronserter. Uploaded file must be a .txt file type, encapsulated file types like .doc or
.rtf are not supported.
Note that by using this option, only step 3 = intron insertion
is performed, and step 1 (codon optimization), step 2 (cut site removal) and
step 4 (fine tuning) are skipped. This is intentionally done to leave your
input DNA sequence untouched, i.e. to assure that your input DNA sequence,
which might have been optimized by other means, e. g. for optimal mRNA
structure, is not altered by Intronserter, except for intron insertion.
For an example, click on the corresponding button below.
OUTPUT :: Output
Parameter
| If "--custom--" was chosen, remove additional user defined restriction digest recognition sites |
If "--custom--" was chosen as a restriction digest site to be removed,
use this parameter to supply your restriction digest recogition sites that
should be removed from the DNA sequence. Note that the length of each recognition sequence
must be of either 6 or 8 nt; other lengths are
not
supported. |
| Start region exon length |
Start region exon length for automatic insertion (lesser equals) (see figure 2 above). |
| Target exon length |
Target exon length for automatic insertion (about) (see figure 2 above). |
| Max exon length |
Max exon length for automatic insertion (lesser equals) (see figure 2 above). |
| End region exon length |
End exon length for automatic insertion (lesser equals) (see figure 2 above). |
| Supersede automatic intron insertion process |
This parameter is to force the utilization of a manual intron position list. If activated,
the automatic intron position determination is switched off and the supplied positions are used
instead. This is useful for other species, for instance when only one single intron at the end
of the sequences should be inserted. |
| User defined intron insertion position list |
If manually provided positions should be used instead, forced by the parameter
"Supersede automatic intron insertion process",
the positions defined here are used instead.
Positions have to supplied as a comma-separated list of integer values.
Note that the positions have to be provided in nt coordinates, not in
amino acid coordinates.
Note further that the positions might not be matched exactly, but only approximately,
which is the case when the nucleotide pair, e.g. "G^G", cannot be found at these
positions. |
| Intron sequence to be inserted |
The intron sequence that is inserted into the cDNA sequence.
The default is the first
intron of rbcS2 of
C. reinhardtii
(or briefly, rbcS2i1).
|
| Optional: substitute the last intron for the below defined second sequence |
Use this option to substitute the last intron for a different intron sequence,
such as the second
intron of rbcS2 of
C. reinhardtii
(or briefly, rbcS2i2).
This option is recommend in C-terminal fusions in the pOptimized vector toolkit
(see Baier et al, 2018), and for other vectors (see figure 3 above). |
| If "substitute last intron" was activated, user defined last intron sequence |
Intron DNA sequence of the last intron, if it should be substituted.
The default is the second
intron of rbcS2 of
C. reinhardtii. |
| DNA sequence input - only insert introns |
Use this option to
only insert introns (step 3),
and not do any other optimzation steps.
With this option, codon optimzation (step 1),
cut site removal (step 2) and fine-tuning (step 4) are not performed. |
| User defined nucleotide addition (DNA) |
User defined nucleotide addition of any length that will be introduced at the
5'-end,
such as a restriction digest recognition site, e.g. "GGATCC". |
| Start codon insertion |
Use this option to insert a heading start codon (ATG, amino acid M)
at the 5'-end
of the sequence. |
| Linker peptide (amino acid sequence) |
User defined amino acid sequence of any length that will be introduced
at the 5'-end,
such as the peptide linker "GSGS".
Note that the additionally introduced amino acids are considered during the intron insertion
process. |
| Start codon removal |
Use this option to remove the native "start codon" (M, Met) of the input
amino acid sequence. |
| Remove stop codon |
Use this option to remove the native "stop codon" (*) of the input
amino acid sequence.
Note that this option is automatically activated if you request a linker peptide at the 3'-end,
in order to avoid accidental premature translation termination. |
| Linker peptide (amino acid sequence) |
User defined amino acid sequence of any length that will be appended
to the 3'-end,
such as the peptide linker "GSGS".
Note that the additionally introduced amino acids are considered during the intron insertion
process. |
| Stop codon insertion |
Use this option to append a trailing stop codon (TAA, TGA or TAG, amino acid *)
at the 3'-end
of the sequence.
Note that this option is ignored if you request a linker peptide at the 3'-end,
in order to avoid accidental premature translation termination. |
| User defined nucleotide addition (DNA) |
User defined nucleotide addition of any length that will be appended to the
3'-end,
such as a restriction digest recognition site, e.g. "GGATCC". |
| Codon optimize using codon usage table for |
Choose one of the two build-in codon usage tables for
C. reinhardtii.
Recommended is the Kazusa codon usage table.
If you want to supply your own codon usage table, for instance if you are working
with another species or manually derived a codon usage table from, e.g. the top 1000 expressed
genes from C. reinhardtii,
you have to choose this directly from the beginning by using the second option.
|
| Remove the following restriction digest sites from back-translated sequence |
All restriction digest sites that are checked are avoided in the
optimized sequence. If your restriction digest site is not available in the build-in
options, you can activate the option "--custom--" and supply your restriction digest sites
to be avoided in the text area field below. |
| Nucleotide pair between which introns are inserted (default G^G) |
The nucleotide pair where introns are inserted, by default between "G^G". |
| Restriction digest site for subsequent cloning |
The restriction digest site that will be inserted at the
5'-end.
If a restriction digest site is not wanted, choose the option "--None--".
Use the option "--custom--" if you want to supply your own nucleotide addition. |
| Restriction digest site for subsequent cloning |
The restriction digest site that will be appended to the
3'-end.
If a restriction digest site is not wanted, choose the option "--None--".
Use the option "--custom--" if you want to supply your own nucleotide addition. |
|
|
