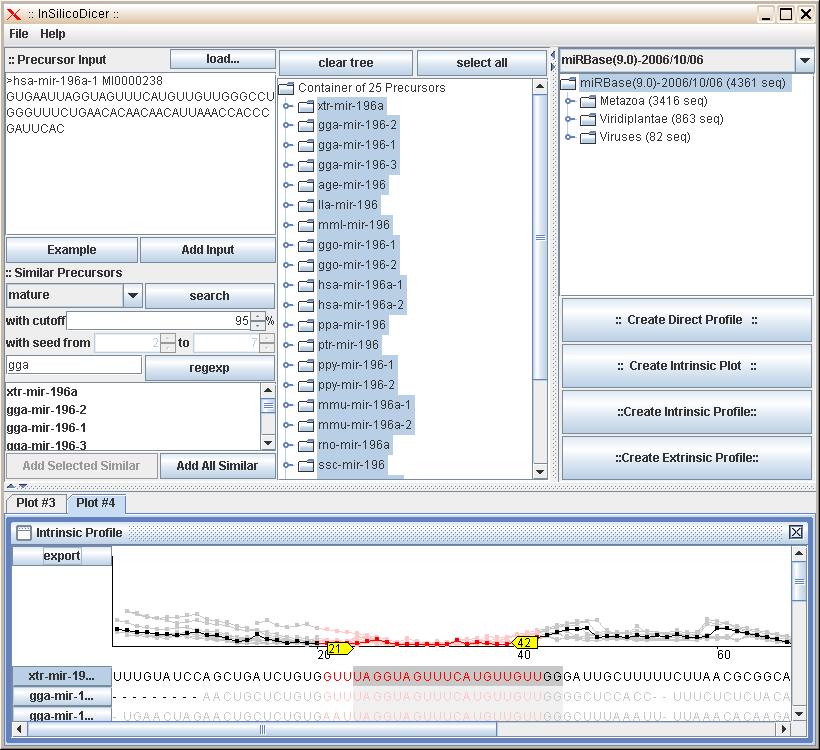
The figure below shows a screenshot of the InSilicoDicer program. The main screen is divided into several regions,
one for each part of the program you can use. Please move the mouse cursor over the different regions of the screenshot to
display a short summary of the parts' function. Clicking on a region will lead you to a more detailed description of that
regions' functionality.
��
Input
InSilicoDicer accepts one or more sequences in FASTA format as input. The data can be loaded from local disc storage, inserted
via cut&paste into the textarea field from another program or copied via drag&drop from the miRBase area.
The following example shows a sequence in the accepted style of the FASTA format:
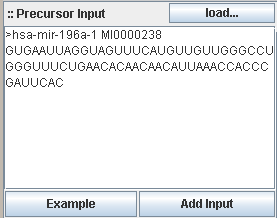 Each sequence in FASTA format starts with a '>' character. The following word is the (mandatory!) sequence id, the rest of
the first line is the sequence description (optional). The next lines up to the occurrence of another '&' character at
the beginning of a line contain sequence information. Any nucleotide following the reduced IUPAC alphabet for nucleotides is
supported. Any character not matching A|C|G|T|U|N is interpreted as aNy character.
Each sequence in FASTA format starts with a '>' character. The following word is the (mandatory!) sequence id, the rest of
the first line is the sequence description (optional). The next lines up to the occurrence of another '&' character at
the beginning of a line contain sequence information. Any nucleotide following the reduced IUPAC alphabet for nucleotides is
supported. Any character not matching A|C|G|T|U|N is interpreted as aNy character.
��
Search
InSilicodicer offers a variety of possibilities to match your inserted sequence data against choosen
database sequence(s). The algorithm used for the local and global alignments is a Smith-Waterman
implementation.
Searching against complete sequences, known mature regions or seed regions with a cut-off limitation (in percent, default is
95% similarity) is supported. The search result can be restricted using a regular expression in Java style (see Java Regular
Expressions for more information about this notation).
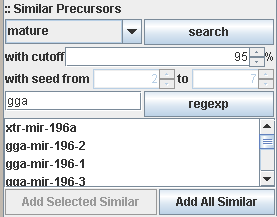 For example, in the above image, a click on the regexp button will remove all results from the list which do not start with
gga (gga is implicitly extended to the regular expression gga.*)
For example, in the above image, a click on the regexp button will remove all results from the list which do not start with
gga (gga is implicitly extended to the regular expression gga.*)
Either all found search results or only a selected variety from the list can be copied to the container area for further
processing.
��
container
The precursor container area holds all precursors you collected by inserting your sequence data and/or by one or more searches.
All selected precursors are used for further processing.
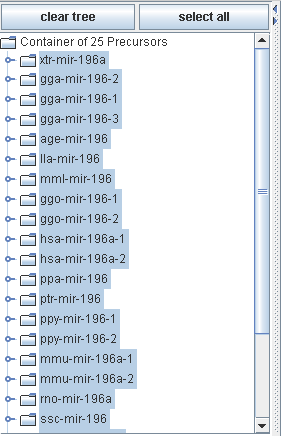 All precursor elements are organized in a tree like structure. Buttons for clearing the tree and selecting all precursors are supported.
��
All precursor elements are organized in a tree like structure. Buttons for clearing the tree and selecting all precursors are supported.
��
database
InSilicoDicer comes with different versions of the miRBase. By default the newest version is selected. The user can choose the version of
the miRBase and - much more important - can select the species he is interested in. Additionally, single sequences can be copied to the
input field by using drag&drop.
 Different databases can be choosen with the upper Drop-Down list. Currently (January 2007) all miRBase databases from version 7.0 to
version 9.0 are available. The database is represented in a tree like structure separated into species and subspecies. Multiselection
is allowed.
��
Different databases can be choosen with the upper Drop-Down list. Currently (January 2007) all miRBase databases from version 7.0 to
version 9.0 are available. The database is represented in a tree like structure separated into species and subspecies. Multiselection
is allowed.
��
action
InSilicoDicer currently supports four different methods (called actions) to
predict the mature region of the input sequence(s). These can be chosen by pressing
the respective button in the action area.
The intrinsic plot, intrinsic profile and extrinisic profile actions have not changed since the initial version of InSilicoDicer, for a detailed explanation of the algorithm used in these actions please have a look at the Bachelor thesis of Sylvia Tippmann.
The direct profile action aligns a set of mircoRNA's with known mature regions against the unknown input sequence(s). The result - a couple of local alignments - is clustered. The best cluster (containing the most overlapping sequences for example) is used to predict the mature region of the unknown sequence.
The results of all actions are displayed in the result area of InSilicDicer.
result(s)
All results are shown as interactive graphical plot. The user can export the result as text (for further processing in another program), as image in a common bitmap format (jpg,png - for web/slide presentation) or as scalable vector graphic (SVG - for high quality prints).
