ChromA - Welcome
Chromatogram Alignment for Chromatography-Mass
Spectrometry is a web based tool for the alignment and
visualization of data from Chromatography-Mass Spectrometry.
Please note that ChromA is now part of Maltcms. The version on this server
is not the most recent one and will not receive further
functional updates. Please consider using Maltcms with the embedded
chroma.properties pipeline instead!
Consider the case of a Metabolomics experiment, where you have
measured the response of an organism's metabolome under different
conditions. Now, you want to compare your data, noting, that some
peaks seem to be conserved, while other peaks of known compounds
are shifted in retention time between experiments. This is
especially evident if you compare replicate experiments, let
alone the intensity variation, the retention time shifts behave
non-linearly and increase your work-load in assigning peaks
between chromatograms.
So what you have will look comparable to the unaligned data in
the upper part of the following image (Click to view complete
TICs):
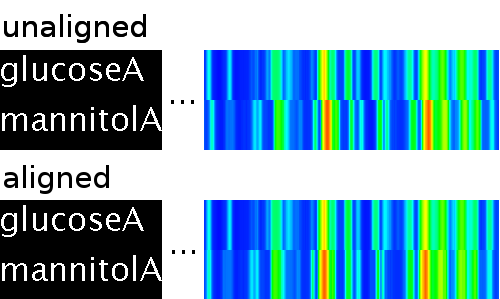
What you get using the ChromA
webinterface or the Web Start version
is shown in the lower part of the image. Please note,
that the largest nonlinear distortions usually occur towards the
middle of the chromatograms. Thus, the preview only shows a
section of the original TIC and the aligned TIC in the middle of
the original chromatograms.
|