Authors: R. Wittler, M. Smith
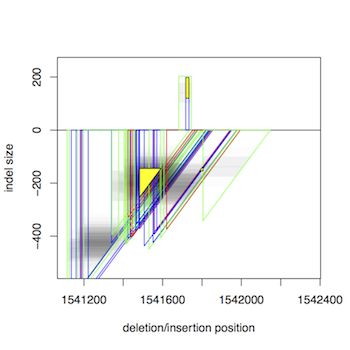 This tool to detect structural variations is specifically designed to cluster
short-read paired-end data into possibly overlapping predictions for deletions
and insertions. The method does not make any assumptions on the composition
of the data, such as the number of samples, heterogeneity, polyploidy, etc.
Taking paired ends mapped to a reference genome as input, it iteratively
merges mappings to clusters based on a similarity score that takes both
the putative location and size of an indel into account.
This tool to detect structural variations is specifically designed to cluster
short-read paired-end data into possibly overlapping predictions for deletions
and insertions. The method does not make any assumptions on the composition
of the data, such as the number of samples, heterogeneity, polyploidy, etc.
Taking paired ends mapped to a reference genome as input, it iteratively
merges mappings to clusters based on a similarity score that takes both
the putative location and size of an indel into account.
|
|
