
|
 |

| |
|
|
|
|
The problem of RNA secondary structure prediction is the following: given the primary sequence of an RNA molecule (a string over the alphabet A, C, G, U), find the "best" set of pairs that the bases (characters) can form within this string. Bases can also remain unpaired.
This gives us two challenges:
- find a combinatorial way to enumerate all possible sets of base pairs for the presented optimization problem. Remember, valid pairs are
A-U, C-G, G-U and vice versa.
- find a biological relevant definition for "best", i.e. define the optimization function.
|
|
|
In general, pairs that cross each other, e.g. A pairs with U and C with G in the sequence ACUG, lead to an NP-hard problem, which can not be solved in affordable compute time. Thus, crossing base pairs (often called pseudoknots) are excluded from most RNA algorithms. Following this restraint, all base pairs are either adjacent to or nested within each other.
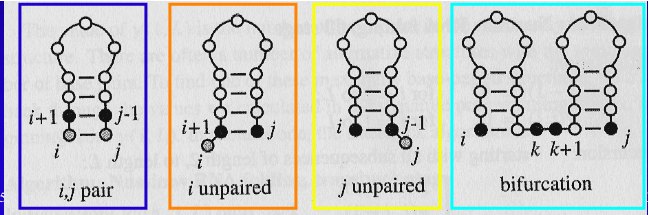
The Nussinov algorithm is using four possible ways to form a RNA structure for a subsequence i, j:
- pair: add a new pair
i, j onto the structure found for subsequence i+1, j-1
i unpaired: add unpaired position i onto structure for subsequence i+1, jj unpaired: add unpaired position j onto structure for subsequence i, j-1- bifurcation: combine two substructures
i, k and k+1, j
|
 |
|
Three definitions for the optimization function
- base pair maximization: in 1978, Ruth Nussinov defined the best secondary structure as the one with the maximum number of valid base pairs. Her dynamic programming approach can find the best structure within O(n3) time and O(n2) space. Although this criterion is too simplistic to give accurate structure predictions, the example is instructive because the mechanics of the algorithm carry over to more sophisticated algorithms.
- Minimum free energy (MFE): Since stability of an RNA structure comes from stacking energies, not from the hydrogen bonds formed between base pairs, the number of base pairs is less important than the types of stacked base pairs. Their energy contributions have been measured in elaborate wet lab experiments for small building blocks of RNA structures (thermodynamic parameters). All different secondary structures for the same sequence are then scored with the sum of all their building block energies. The one with the lowest, i.e. the best, free energy is the winner. Since only the optimization criterion changes, this kind of prediction can also be done in O(n3) time and O(n2) space. A popular implementation is RNAfold.
Unfortunately, the biological correct structure is still not found as the MFE structure, because of base modifications, structure changing protein interactions, insufficient thermodynamic parameters and many more reasons. Thus, it is absolutely necessary to also look at structures with slightly worse MFE values, called suboptimals. RNAsubopt can enumerate them.
- MFE Shapes: The number of different secondary structures heavily grows with sequence length, but they often differ from each other by only tiny structural rearrangements like addition or removal of a base pair, or a slight shift in position of a small bulge loop. Structures can be pooled according to their abstract shape. Generally, an abstract shape gives information about the arrangement of structural elements such as stacked regions, but no concrete base pairs. The MFE structure within each shape class is called "shrep".
RNAsubopt reports confusingly long list of different structures. With the abstraction technique, implemented in RNAshapes, we can directly focus on important differences.
A very popular representation of RNA secondary structures is the " Vienna Dot Bracket String", where an unpaired base is represented by a single dot . and the partners of a base pair as a bracket ().
A very small clover leaf might look like ((.((..)).(((.)))((...))))A nice tool for the visualization of many RNA secondary structures is RNAmovies. It renders a sequential animation from individual structures and thus allows for a convenient overview of many alternative structures of one RNA molecule.
|
|









