RapidShapes computes a thermodynamic matcher (TDM), using a runtime heuristic for probabilistic shape analysis.
It uses a pipeline to calculate the exact shape probabilities.
This pipeline contains the following three steps:
1. Creates a set of the most likely shapes.
2. Translates the Shape-Strings into ADP-grammars.
3. Each ADP-grammar is then translated into a TDM in form of a C++ program.
The gain in runtime grows with the sequence length using the RapidShape heuristic. Thus, we are the first time able to analyse sequences with more then 400 nucleotides or more.
Depending on the alpha-threshold, RapidShapes clearly speeds up shape analysis compared with the previous methods.
Thus, probabilistic shape analysis has become feasible in medium-scale applications, such as the screening of RNA transcripts in a bacterial genome.
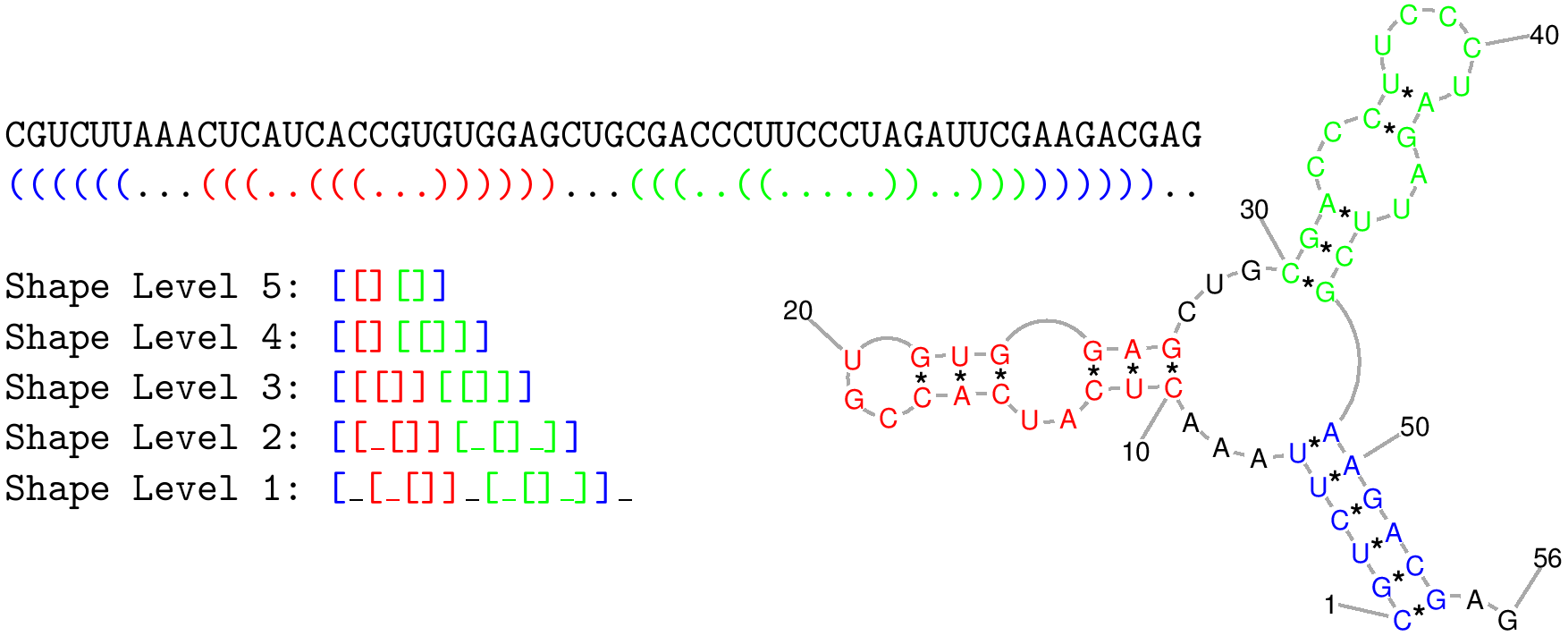
Figure 1: Shows a RNA shape and the different levels of abstract representation. The abstract shape classes have a huge advantage: many similar RNA shapes can be combined to an abstract shape class. The abstraction level is defined by numbers, where 5 is the most abstract level and 1 is the least abstract level. RapidShapes use this abstraction for generating shape specific thermodynamic matcher.
