The BiBiServ team discontinued the online service for InSilicoDicer. Please contact the author(s)
of InsilicoDicer for any questions concerning InSilicoDicer.
MicroRNAs are short RNA sequences that use antisense complementarity to repress expression of specific messenger RNAs.
If we could fully understand their biogenesis and function in vivo, we would possibly be able to predict a many more
target genes and to annotate them and furthermore knock down unwanted, maybe pathological, ones through introduction
of artificial miRNA genes. The maturation of these smallRNAs, however, appears to be of high complexity since a large
number of proteins are involved and, dependent on the organism, diverse procedures seem to apply. Ever so much
bioinformatical research has been done developing different approaches to reconstruct the miRNA pathway, and was partially
successful already. Some programs were published to predict possible miRNA precursors from whole genomes, and there exist
algorithms which match the mature miRNA with its corresponding target gene. Unfortunately, little is known about the step
in between, the processing of precursors to mature and functional active miRNA, so there are only few approaches to close
this prediction gap. According to one of the central miRNA processing enzymes the tool, that tries to bridge the gap, is
named "In-Silico-Dicer".
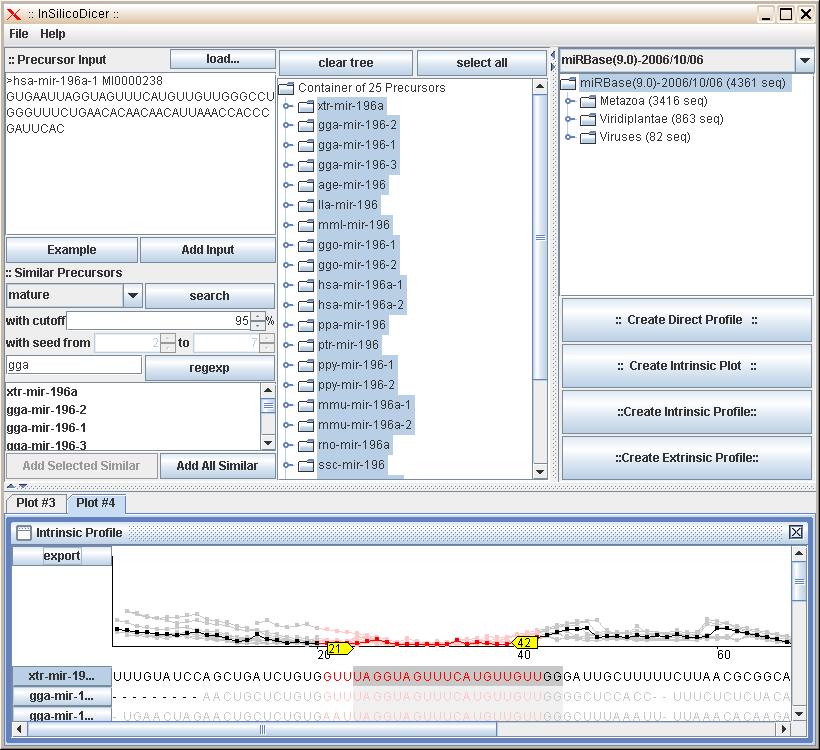
For the prediction of mature miRNA from known precursors In-Silico-Dicer deploys two different approaches which can be used
seperately. For algorithmic details of the approaches please have a look at references.
